PyNN 0.8 beta 1 release notes¶
November 15th 2013
Welcome to the first beta release of PyNN 0.8!
For full information about what’s new in PyNN 0.8, see the PyNN 0.8 alpha 1 release notes.
Brian backend¶
The main new feature in this beta release is the reappearance of the Brian backend, updated to work with the 0.8 API. You will need version 1.4 of Brian. There are still some rough edges, but we encourage anyone who has used Brian with PyNN 0.7 to try updating your scripts now, and give it a try.
New and improved connectors¶
The library of Connector classes has been extended. The
DistanceDependentProbabilityConnector (DDPC) has been generalized, resulting
in the IndexBasedProbabilityConnector, with which the connection
probability can be specified as any function of the indices i and j of the
pre- and post-synaptic neurons within their populations. In addition, the
distance expression for the DDPC can now be a callable object (such as a
function) as well as a string expression.
The ArrayConnector allows connections to be specified as an explicit
boolean matrix, with shape (m, n) where m is the size of the presynaptic
population and n that of the postsynaptic population.
The CSAConnector has been updated to work with the 0.8 API.
The FromListConnector and FromFileConnector now support
specifying any synaptic parameter (e.g. parameters of the synaptic plasticity
rule), not just weight and delay.
API changes¶
The set() function now matches the Population.set() method,
i.e. it takes one or more parameter name/value pairs as keyword arguments.
Two new functions for advancing a simulation have been added: run_for()
and run_until(). run_for() is just an alias for run().
run_until() allows you to specify the absolute time at which a
simulation should stop, rather than the increment of time. In addition, it is
now possible to specify a call-back function that should be called at
intervals during a run, e.g.:
>>> def report_time(t):
... print("The time is %g" % t)
... return t + 100.0
>>> run_until(300.0, callbacks=[report_time])
The time is 0
The time is 100
The time is 200
The time is 300
One potential use of this feature is to record synaptic weights during a simulation with synaptic plasticity.
We have changed the parameterization of STDP models. The A_plus and A_minus parameters have been moved from the weight-dependence components to the timing-dependence components, since effectively they describe the shape of the STDP curve independently of how the weight change depends on the current weight.
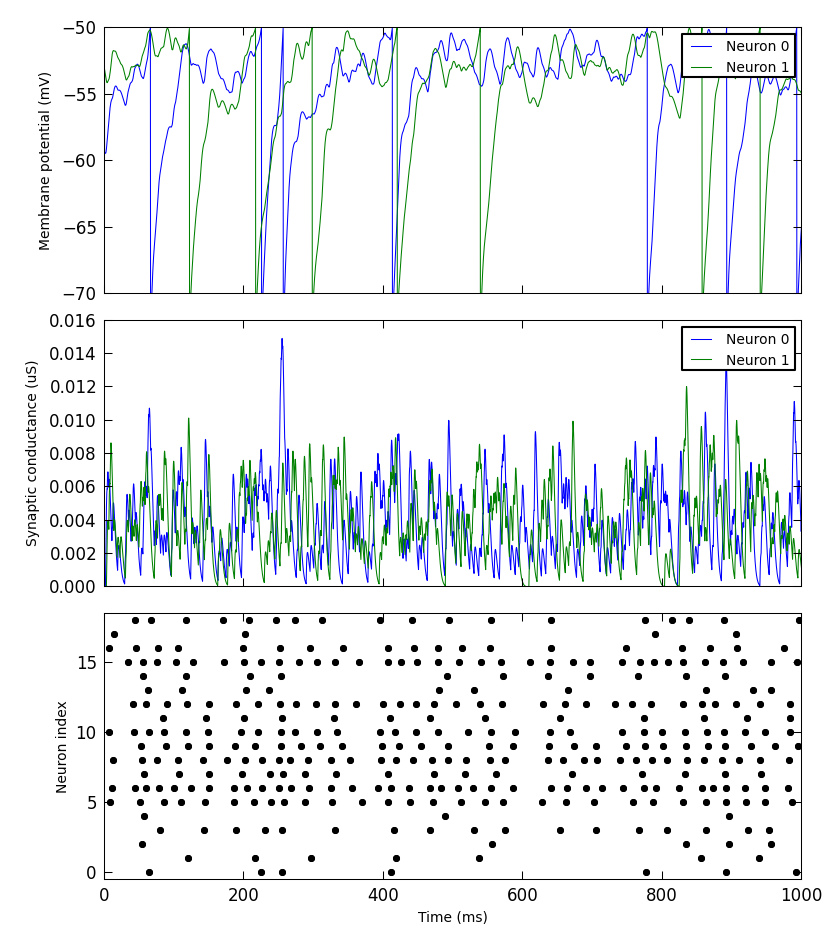
Simple plotting¶
We have added a small library to make it simple to produce simple plots of data recorded from a PyNN simulation. This is not intended for publication-quality or highly-customized plots, but for basic visualization.
For example:
from pyNN.utility.plotting import Figure, Panel
...
population.record('spikes')
population[0:2].record(('v', 'gsyn_exc'))
...
data = population.get_data().segments[0]
vm = data.filter(name="v")[0]
gsyn = data.filter(name="gsyn_exc")[0]
Figure(
Panel(vm, ylabel="Membrane potential (mV)"),
Panel(gsyn, ylabel="Synaptic conductance (uS)"),
Panel(data.spiketrains, xlabel="Time (ms)", xticks=True)
).save("simulation_results.png")
Gap junctions¶
The NEURON backend now supports gap junctions. This is not yet an official part of the PyNN API, since any official feature must be supported by at least two backends, but could be very useful to modellers using NEURON.
Other changes¶
The default precision for the NEST backend has been changed to “off_grid”. This reflects the PyNN philosophy that defaults should prioritize accuracy and compatibility over performance. (We think performance is very important, it’s just that any decision to risk compromising accurary or interoperability should be made deliberately by the end user.)
The Izhikevich neuron model is now available for the NEURON, NEST and Brian backends, although there are still some problems with injecting current when using the NEURON backend.
A whole bunch of bugs have been fixed: see the issue tracker.
