# encoding: utf8
"""
A very simple example of using STDP.
A single postsynaptic neuron fires at a constant rate. We connect several
presynaptic neurons to it, each of which fires spikes with a fixed time
lag or time advance with respect to the postsynaptic neuron.
The weights of these connections are very small, so they will not
significantly affect the firing times of the post-synaptic neuron.
We plot the amount of potentiation or depression of each synapse as a
function of the time difference.
Usage: python simple_STDP.py [-h] [--plot-figure] [--debug DEBUG] simulator
positional arguments:
simulator neuron, nest, brian or another backend simulator
optional arguments:
-h, --help show this help message and exit
--plot-figure Plot the simulation results to a file
--fit-curve Calculate the best-fit curve to the weight-delta_t measurements
--debug DEBUG Print debugging information
"""
from __future__ import division
from math import exp
import numpy
import neo
from quantities import ms
from pyNN.utility import get_simulator, init_logging, normalized_filename
from pyNN.utility.plotting import DataTable
from pyNN.parameters import Sequence
# === Parameters ============================================================
firing_period = 100.0 # (ms) interval between spikes
cell_parameters = {
"tau_m": 10.0, # (ms)
"v_thresh": -50.0, # (mV)
"v_reset": -60.0, # (mV)
"v_rest": -60.0, # (mV)
"cm": 1.0, # (nF)
"tau_refrac": firing_period / 2, # (ms) long refractory period to prevent bursting
}
n = 60 # number of synapses / number of presynaptic neurons
delta_t = 1.0 # (ms) time difference between the firing times of neighbouring neurons
t_stop = 10 * firing_period + n * delta_t
delay = 3.0 # (ms) synaptic time delay
# === Configure the simulator ===============================================
sim, options = get_simulator(("--plot-figure", "Plot the simulation results to a file", {"action": "store_true"}),
("--fit-curve", "Calculate the best-fit curve to the weight-delta_t measurements", {"action": "store_true"}),
("--dendritic-delay-fraction", "What fraction of the total transmission delay is due to dendritic propagation", {"default": 1}),
("--debug", "Print debugging information"))
if options.debug:
init_logging(None, debug=True)
sim.setup(timestep=0.01, min_delay=delay, max_delay=delay)
# === Build the network =====================================================
def build_spike_sequences(period, duration, n, delta_t):
"""
Return a spike time generator for `n` neurons (spike sources), where
all neurons fire with the same period, but neighbouring neurons have a relative
firing time difference of `delta_t`.
"""
def spike_time_gen(i):
"""Spike time generator. `i` should be an array of indices."""
return [Sequence(numpy.arange(period + j * delta_t, duration, period)) for j in (i - n // 2)]
return spike_time_gen
spike_sequence_generator = build_spike_sequences(firing_period, t_stop, n, delta_t)
# presynaptic population
p1 = sim.Population(n, sim.SpikeSourceArray(spike_times=spike_sequence_generator),
label="presynaptic")
# single postsynaptic neuron
p2 = sim.Population(1, sim.IF_cond_exp(**cell_parameters),
initial_values={"v": cell_parameters["v_reset"]}, label="postsynaptic")
# drive to the postsynaptic neuron, ensuring it fires at exact multiples of the firing period
p3 = sim.Population(1, sim.SpikeSourceArray(spike_times=numpy.arange(firing_period - delay, t_stop, firing_period)),
label="driver")
# we set the initial weights to be very small, to avoid perturbing the firing times of the
# postsynaptic neurons
stdp_model = sim.STDPMechanism(
timing_dependence=sim.SpikePairRule(tau_plus=20.0, tau_minus=20.0,
A_plus=0.01, A_minus=0.012),
weight_dependence=sim.AdditiveWeightDependence(w_min=0, w_max=0.0000001),
weight=0.00000005,
delay=delay,
dendritic_delay_fraction=float(options.dendritic_delay_fraction))
connections = sim.Projection(p1, p2, sim.AllToAllConnector(), stdp_model)
# the connection weight from the driver neuron is very strong, to ensure the
# postsynaptic neuron fires at the correct times
driver_connection = sim.Projection(p3, p2, sim.OneToOneConnector(),
sim.StaticSynapse(weight=10.0, delay=delay))
# == Instrument the network =================================================
p1.record('spikes')
p2.record(['spikes', 'v'])
class WeightRecorder(object):
"""
Recording of weights is not yet built in to PyNN, so therefore we need
to construct a callback object, which reads the current weights from
the projection at regular intervals.
"""
def __init__(self, sampling_interval, projection):
self.interval = sampling_interval
self.projection = projection
self._weights = []
def __call__(self, t):
self._weights.append(self.projection.get('weight', format='list', with_address=False))
return t + self.interval
def get_weights(self):
return neo.AnalogSignalArray(self._weights, units='nA', sampling_period=self.interval * ms,
channel_index=numpy.arange(len(self._weights[0])),
name="weight")
weight_recorder = WeightRecorder(sampling_interval=1.0, projection=connections)
# === Run the simulation =====================================================
sim.run(t_stop, callbacks=[weight_recorder])
# === Save the results, optionally plot a figure =============================
filename = normalized_filename("Results", "simple_stdp", "pkl", options.simulator)
p2.write_data(filename, annotations={'script_name': __file__})
presynaptic_data = p1.get_data().segments[0]
postsynaptic_data = p2.get_data().segments[0]
print("Post-synaptic spike times: %s" % postsynaptic_data.spiketrains[0])
weights = weight_recorder.get_weights()
final_weights = numpy.array(weights[-1])
deltas = delta_t * numpy.arange(n // 2, -n // 2, -1)
print("Final weights: %s" % final_weights)
plasticity_data = DataTable(deltas, final_weights)
if options.fit_curve:
def double_exponential(t, t0, w0, wp, wn, tau):
return w0 + numpy.where(t >= t0, wp * numpy.exp(-(t - t0) / tau), wn * numpy.exp((t - t0) / tau))
p0 = (-1.0, 5e-8, 1e-8, -1.2e-8, 20.0)
popt, pcov = plasticity_data.fit_curve(double_exponential, p0, ftol=1e-10)
print("Best fit parameters: t0={0}, w0={1}, wp={2}, wn={3}, tau={4}".format(*popt))
if options.plot_figure:
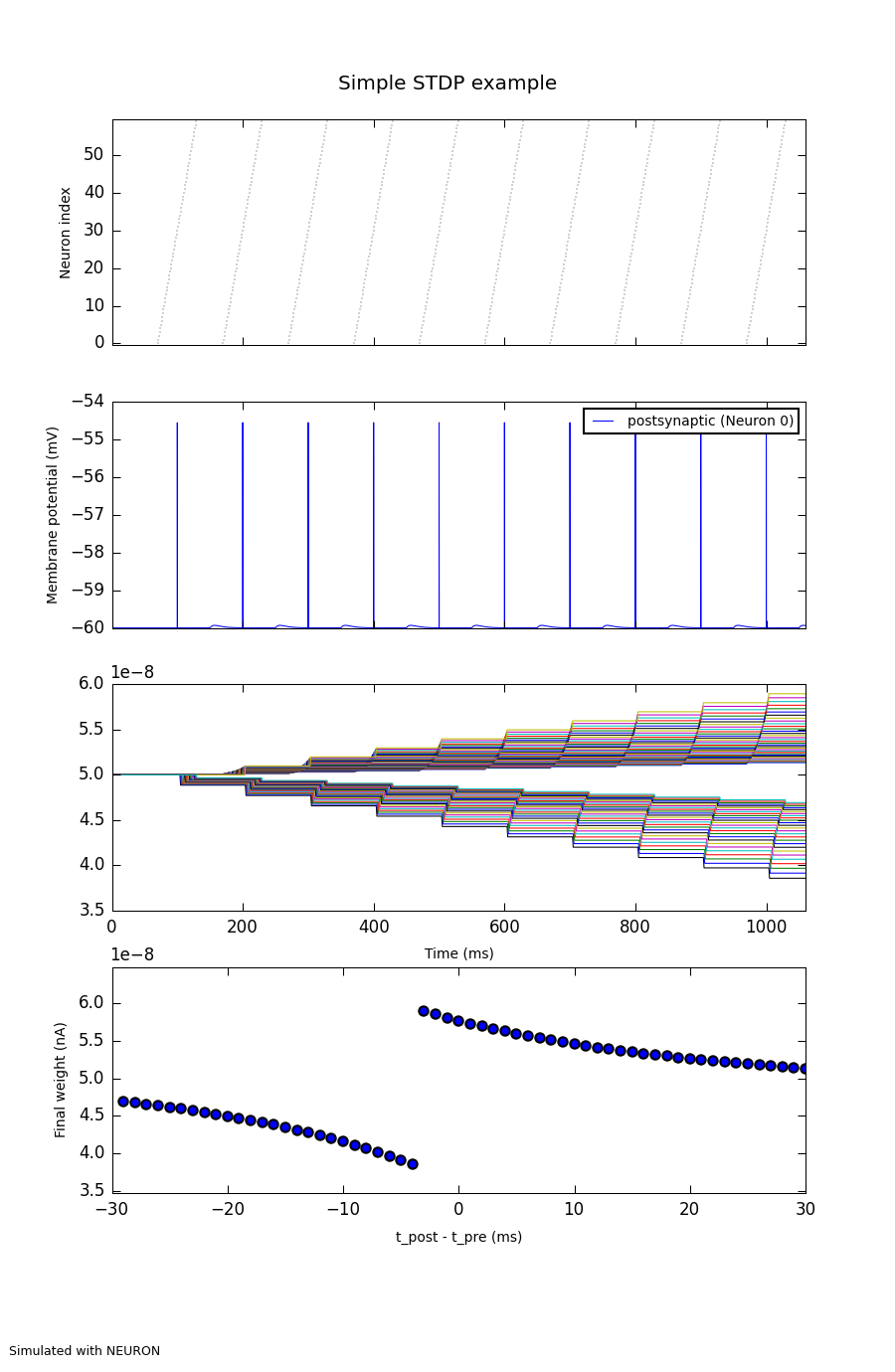
from pyNN.utility.plotting import Figure, Panel, DataTable
figure_filename = filename.replace("pkl", "png")
Figure(
# raster plot of the presynaptic neuron spike times
Panel(presynaptic_data.spiketrains,
yticks=True, markersize=0.2, xlim=(0, t_stop)),
# membrane potential of the postsynaptic neuron
Panel(postsynaptic_data.filter(name='v')[0],
ylabel="Membrane potential (mV)",
data_labels=[p2.label], yticks=True, xlim=(0, t_stop)),
# evolution of the synaptic weights with time
Panel(weights, xticks=True, yticks=True, xlabel="Time (ms)",
legend=False, xlim=(0, t_stop)),
# scatterplot of the final weight of each synapse against the relative
# timing of pre- and postsynaptic spikes for that synapse
Panel(plasticity_data,
xticks=True, yticks=True, xlim=(-n / 2 * delta_t, n / 2 * delta_t),
ylim=(0.9 * final_weights.min(), 1.1 * final_weights.max()),
xlabel="t_post - t_pre (ms)", ylabel="Final weight (nA)",
show_fit=options.fit_curve),
title="Simple STDP example",
annotations="Simulated with %s" % options.simulator.upper()
).save(figure_filename)
print(figure_filename)
# === Clean up and quit ========================================================
sim.end()