PyNN 0.8.0 release notes¶
October 5th 2015
Welcome to the final release of PyNN 0.8.0!
For PyNN 0.8 we have taken the opportunity to make significant, backward-incompatible changes to the API. The aim was fourfold:
to simplify the API, making it more consistent and easier to remember;
to make the API more powerful, so more complex models can be expressed with less code;
to allow a number of internal simplifications so it is easier for new developers to contribute;
to prepare for planned future extensions, notably support for multi-compartmental models.
We summarize here the main changes between versions 0.7 and 0.8 of the API.
Creating populations¶
In previous versions of PyNN, the Population constructor was called
with the population size, a BaseCellType sub-class such as
IF_cond_exp and a dictionary of parameter values. For example:
p = Population(1000, IF_cond_exp, {'tau_m': 12.0, 'cm': 0.8}) # PyNN 0.7
This dictionary was passed to the cell-type class constructor within the
Population constructor to create a cell-type instance.
The reason for doing this was that in early versions of PyNN, use of native
NEST models was supported by passing a string, the model name, as the cell-type
argument. Since PyNN 0.7, however, native models have been supported with the
NativeCellType class, and passing a string is no longer allowed.
It makes more sense, therefore, for the cell-type instance to be created by the
user, and to pass a cell-type instance, rather than a cell-type class, to the
Population constructor.
There is also a second change: specification of parameters for cell-type classes is now done via keyword arguments rather than a single parameter dictionary. This is for consistency with current sources and synaptic plasticity models, which already use keyword arguments.
The example above should be rewritten as:
p = Population(1000, IF_cond_exp(tau_m=12.0, cm=0.8)) # PyNN 0.8
or:
p = Population(1000, IF_cond_exp(**{'tau_m': 12.0, 'cm': 0.8})) # PyNN 0.8
or:
cell_type = IF_cond_exp(tau_m=12.0, cm=0.8) # PyNN 0.8
p = Population(1000, cell_type)
The first form, with a separate parameter dictionary, is still supported for the time being, but is deprecated and may be removed in future versions.
Specifying heterogeneous parameter values¶
In previous versions of PyNN, the Population constructor supported
setting parameters to either homogeneous values (all cells in the population
have the same value) or random values. After construction, it was possible to
change parameters using the Population.set(), Population.tset()
(for topographic set - parameters were set by using an array of the same
size as the population) and Population.rset() (for random set) methods.
In PyNN 0.8, setting parameters is simpler and more consistent, in that both
when constructing a cell type for use in the Population constructor
(see above) and in the Population.set() method, parameter values can be
any of the following:
a single number - sets the same value for all cells in the
Population;a
RandomDistributionobject - for each cell, sets a different random value drawn from the distribution;a list or 1D NumPy array of the same size as the
Population;a function that takes a single integer argument; this function will be called with the index of every cell in the
Populationto return the parameter value for that cell.
See Model parameters and initial values for more details and examples.
The call signature of the Population.set() method has also been changed;
now parameters should be specified as keyword arguments. For example, instead
of:
p.set({"tau_m": 20.0}) # PyNN 0.7
use:
p.set(tau_m=20.0) # PyNN 0.8
and instead of:
p.set({"tau_m": 20.0, "v_rest": -65}) # PyNN 0.7
use:
p.set(tau_m=20.0, v_rest=-65) # PyNN 0.8
Now that Population.set() accepts random distributions and arrays as
arguments, the Population.tset() and Population.rset() methods are
superfluous. As of version 0.8, their use is deprecated and they will
be removed in the next version of PyNN. Their use can be replaced as follows:
p.tset("i_offset", arr) # PyNN 0.7
p.set(i_offset=arr) # PyNN 0.8
p.rset("tau_m", rand_distr) # PyNN 0.7
p.set(tau_m=rand_distr) # PyNN 0.8
Setting spike times¶
Where a single parameter value is already an array, e.g. spike times, this
should be wrapped by a Sequence object. For example, to generate
a different Poisson spike train for every neuron in a population of
SpikeSourceArrays:
def generate_spike_times(i_range):
return [Sequence(numpy.add.accumulate(numpy.random.exponential(10.0, size=10)))
for i in i_range]
p = sim.Population(30, sim.SpikeSourceArray(spike_times=generate_spike_times))
Standardization of random distributions¶
Since its earliest versions PyNN has supported swapping in and out different random number
generators, but until now there has been no standardization of these RNGs;
for example the GSL random number library uses “gaussian” where NumPy uses “normal”.
This limited the usefulness of this feature, especially for the NativeRNG class,
which signals that random number generation should be passed down to the simulator backend
rather than being performed at the Python level.
This has now been fixed. The names of random number distributions and of their parameters have now been standardized, based for the most part on the nomenclature used by Wikipedia. A quick example:
from pyNN.random import NumpyRNG, GSLRNG, RandomDistribution
rd1 = RandomDistribution('normal' mu=0.5, sigma=0.1, rng=NumpyRNG(seed=922843))
rd2 = RandomDistribution('normal' mu=0.5, sigma=0.1, rng=GSLRNG(seed=426482))
Recording¶
Previous versions of PyNN had three methods for recording from populations of
neurons: record(), record_v() and record_gsyn(), for
recording spikes, membrane potentials, and synaptic conductances, respectively.
There was no official way to record any other state variables, for example the
w variable from the adaptive-exponential integrate-and-fire model, or when
using native, non-standard models, although there were workarounds.
In PyNN 0.8, we have replaced these three methods with a single record()
method, which takes the variable to record as its first argument, e.g.:
p.record() # PyNN 0.7
p.record_v()
p.record_gsyn()
becomes:
p.record('spikes') # PyNN 0.8
p.record('v')
p.record(['gsyn_exc', 'gsyn_inh'])
Note that (1) you can now choose to record the excitatory and inhibitory
synaptic conductances separately, (2) you can give a list of variables to
record. For example, you can record all the variables for the
EIF_cond_exp_isfa_ista model in a single command using:
p.record(['spikes', 'v', 'w', 'gsyn_exc', 'gsyn_inh']) # PyNN 0.8
Note that the record_v() and record_gsyn() methods still exist,
but their use is deprecated, and they will be removed in the next version of
PyNN.
A further change is that Population.record() has an optional sampling_interval argument,
allowing recording at intervals larger than the integration time step.
See Recording spikes and state variables for more details.
Retrieving recorded data¶
Perhaps the biggest change in PyNN 0.8 is that handling of recorded data, whether retrieval as Python objects or saving to file, now uses the Neo package, which provides a common Python object model for neurophysiology data (whether real or simulated).
Using Neo provides several advantages:
data objects contain essential metadata, such as units, sampling interval, etc.;
data can be saved to any of the file formats supported by Neo, including HDF5 and Matlab files;
it is easier to handle data when running multiple simulations with the same network (calling
reset()between each one);it is possible to save multiple signals to a single file;
better interoperability with other Python packages using Neo (for data analysis, etc.).
Note that Neo is based on NumPy, and most Neo data objects sub-class the NumPy
ndarray class, so much of your data handling code should work exactly
the same as before.
See Data handling for more details.
Creating connections¶
In previous versions of PyNN, synaptic weights and delays were specified on
creation of the Connector object. If the synaptic weight had its own
dynamics (whether short-term or spike-timing-dependent plasticity), the
parameters for this were specified on creation of a SynapseDynamics
object. In other words, specification of synaptic parameters was split across
two different classes.
SynapseDynamics was also rather complex, and could have both a “fast”
(for short-term synaptic depression and facilitation) and “slow” (for long-term
plasticity) component, although most simulator backends did not support
specifying both fast and slow components at the same time.
In PyNN 0.8, all synaptic parameters including weights and delays are given as
arguments to a SynapseType sub-class such as StaticSynapse or
TsodyksMarkramSynapse. For example, instead of:
prj = Projection(p1, p2, AllToAllConnector(weights=0.05, delays=0.5)) # PyNN 0.7
you should now write:
prj = Projection(p1, p2, AllToAllConnector(), StaticSynapse(weight=0.05, delay=0.5)) # PyNN 0.8
and instead of:
params = {'U': 0.04, 'tau_rec': 100.0, 'tau_facil': 1000.0}
facilitating = SynapseDynamics(fast=TsodyksMarkramMechanism(**params)) # PyNN 0.7
prj = Projection(p1, p2, FixedProbabilityConnector(p_connect=0.1, weights=0.01),
synapse_dynamics=facilitating)
the following:
params = {'U': 0.04, 'tau_rec': 100.0, 'tau_facil': 1000.0, 'weight': 0.01}
facilitating = TsodyksMarkramSynapse(**params) # PyNN 0.8
prj = Projection(p1, p2, FixedProbabilityConnector(p_connect=0.1),
synapse_type=facilitating)
Note that “weights” and “delays” are now “weight” and “delay”. In addition,
the “method” argument to Projection is now called “connector”,
and the “target” argument is now “receptor_type”. The “rng” argument has
been moved from Projection to Connector, and the “space”
argument of Connector has been moved to Projection.
The ability to specify both short-term and long-term plasticity for a given
connection type, in a simulator-independent way, has been removed, although in
practice only the NEURON backend supported this. This functionality will be
reintroduced in PyNN 0.9. If you need this in the meantime, a workaround for the
NEURON backend is to use a NativeSynapseType mechanism - ask on the
mailing list for guidance.
Finally, the parameterization of STDP models has been modified. The A_plus and A_minus parameters have been moved from the weight-dependence components to the timing-dependence components, since effectively they describe the shape of the STDP curve independently of how the weight change depends on the current weight.
Specifying heterogeneous synapse parameters¶
As for neuron parameters, synapse parameter values can now be any of the following:
a single number - sets the same value for all connections in the
Projection;a
RandomDistributionobject - for each connection, sets a different random value drawn from the distribution;a list or 1D NumPy array of the same size as the
Projection(although this is not very useful for random networks, whose size may not be known in advance);a function that takes a single float argument; this function will be called with the distance between the pre- and post-synaptic cell to return the parameter value for that cell.
Accessing, setting and saving properties of synaptic connections¶
In older versions of PyNN, the Projection class had a bunch of methods
for working with synaptic parameters: getWeights(), setWeights(),
randomizeWeights(), printWeights(), getDelays(),
setDelays(), randomizeDelays(), printDelays(),
getSynapseDynamics(), setSynapseDynamics(),
randomizeSynapseDynamics(), and saveConnections().
These have been replace by three methods: get(), set() and
save(). The original methods still exist, but their use is deprecated and
they will be removed in the next version of PyNN. You should update your code
as follows:
prj.getWeights(format='list') # PyNN 0.7
prj.get('weight', format='list', with_address=False) # PyNN 0.8
prj.randomizeDelays(rand_distr) # PyNN 0.7
prj.set(delay=rand_distr) # PyNN 0.8
prj.setSynapseDynamics('tau_rec', 50.0) # PyNN 0.7
prj.set(tau_rec=50.0) # PyNN 0.8
prj.printWeights('exc_weights.txt', format='array') # PyNN 0.7
prj.save('weight', 'exc_weights.txt', format='array') # PyNN 0.8
prj.saveConnections('exc_conn.txt') # PyNN 0.7
prj.save('all', 'exc_conn.txt', format='list') # PyNN 0.8
Also note that all three new methods can operate on several parameters at a time:
weights, delays = prj.getWeights('array'), prj.getDelays('array') # PyNN 0.7
weights, delays = prj.get(['weight', 'delay'], format='array') # PyNN 0.8
prj.randomizeWeights(rand_distr); prj.setDelays(0.4) # PyNN 0.7
prj.set(weight=rand_distr, delay=0.4) # PyNN 0.8
New and improved connectors¶
The library of Connector classes has been extended. The
DistanceDependentProbabilityConnector (DDPC) has been generalized, resulting
in the IndexBasedProbabilityConnector, with which the connection
probability can be specified as any function of the indices i and j of the
pre- and post-synaptic neurons within their populations. In addition, the
distance expression for the DDPC can now be a callable object (such as a
function) as well as a string expression.
The ArrayConnector allows connections to be specified as an explicit
boolean matrix, with shape (m, n) where m is the size of the presynaptic
population and n that of the postsynaptic population.
The CloneConnector takes the connection
matrix from an existing Projection and uses it to create a new Projection,
with the option of changing the weights, delays, receptor type, etc.
The FromListConnector and FromFileConnector now support
specifying any synaptic parameter (e.g. parameters of the synaptic plasticity
rule), not just weight and delay.
The FixedNumberPostConnector now has an option with_replacement,
which controls how the post-synaptic population is sampled,
and affects the incidence of multiple connections between pairs of neurons (“multapses”).
We have added a version of CSAConnector for the NEST backend that passes down the CSA
object to PyNEST’s CGConnect() function.
This greatly speeds up CSAConnector with NEST.
Simulation control¶
Two new functions for advancing a simulation have been added: run_for()
and run_until(). run_for() is just an alias for run().
run_until() allows you to specify the absolute time at which a
simulation should stop, rather than the increment of time. In addition, it is
now possible to specify a call-back function that should be called at
intervals during a run, e.g.:
>>> def report_time(t):
... print("The time is %g" % t)
... return t + 100.0
>>> run_until(300.0, callbacks=[report_time])
The time is 0
The time is 100
The time is 200
The time is 300
One potential use of this feature is to record synaptic weights during a simulation with synaptic plasticity.
The default value of the min_delay argument to setup() is now “auto”,
which means that the simulator should calculate the minimal synaptic delay itself.
This change can lead to large speedups for NEST and NEURON code.
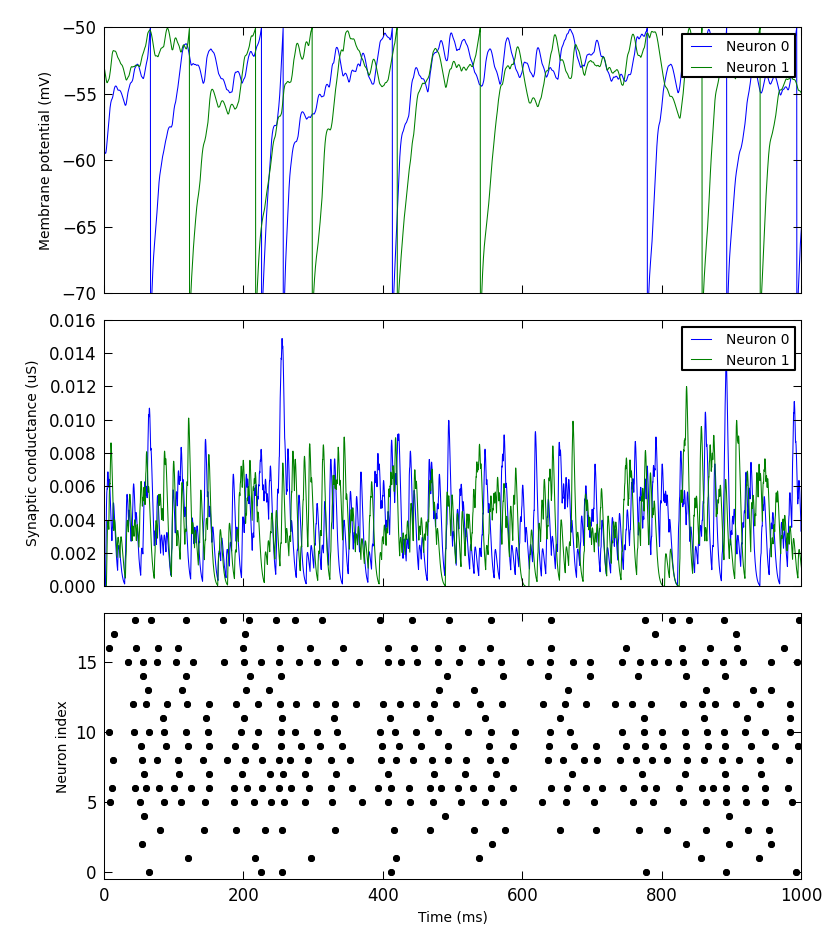
Simple plotting¶
We have added a small library to make it simple to produce simple plots of data recorded from a PyNN simulation. This is not intended for publication-quality or highly-customized plots, but for basic visualization.
For example:
from pyNN.utility.plotting import Figure, Panel
...
population.record('spikes')
population[0:2].record(('v', 'gsyn_exc'))
...
data = population.get_data().segments[0]
vm = data.filter(name="v")[0]
gsyn = data.filter(name="gsyn_exc")[0]
Figure(
Panel(vm, ylabel="Membrane potential (mV)"),
Panel(gsyn, ylabel="Synaptic conductance (uS)"),
Panel(data.spiketrains, xlabel="Time (ms)", xticks=True)
).save("simulation_results.png")
Supported backends¶
PyNN 0.8.0 is compatible with NEST versions 2.6 to 2.8, NEURON versions 7.3 to 7.4, and Brian 1.4. Support for Brian 2 is planned for a future release.
Support for the PCSIM simulator has been dropped since the simulator appears to be no longer actively developed.
The default precision for the NEST backend has been changed to “off_grid”. This reflects the PyNN philosophy that defaults should prioritize accuracy and compatibility over performance. (We think performance is very important, it’s just that any decision to risk compromising accuracy or interoperability should be made deliberately by the end user.)
The Izhikevich neuron model is now available for all backends.
Python compatibility¶
Support for Python 3 has been added (versions 3.3+). Support for Python versions 2.5 and earlier has been dropped.
Changes for developers¶
Other than the internal refactoring, the main change for developers is that we have switched from Subversion to Git. PyNN development now takes place at https://github.com/NeuralEnsemble/PyNN/ We are now taking advantage of the integration of GitHub with TravisCI, to automatically run the test suite whenever changes are pushed to GitHub.
